Stage 7: Lesion analysis
|
Stage summary: Lesion Explorer (LE) and FLEX provide a lesion output with four lesion
classes including periventricular Subcortical Hyperintensity (pvSH), deep white SH (dwSH),
periventricular black holes (pvBH) and dwBH. |
ITK-SNAP keyboard shortcuts
Procedure overview
- Open ITK-SNAP
- Load the T1 {session}_T1, PD {session}_PD_inT1, T2 {session}_T2_inT1 for LE and/or FL {session}_FL_inT1 for FLEX
- Load the lesion mask {session}_LEauto for LE or {session}_FLEXauto for FLEX
- Label lesion using the paintbrush tool
- Save the lesion mask edit {session}_LEedit for LE or {session}_FLEXedit for FLEX
Load images
- Load the T1 {session}_T1, PD {session}_PD_inT1, T2 {session}_T2_inT1``for LE and/or FL ``{session}_FL_inT1 for FLEX
- Adjust intensity
- Turn off crosshairs (x)
- Zoom in to the image until it fills each window (right click and drag with crosshairs tool)
- Adjust centring of axial view if necessary, with navigation tool (may need to do several times during procedure)
- Click on paintbrush tool
Lesion Explorer (LE)
Lesion Explorer (LE) uses PDT2 to perform a semi-automatic lesion analysis.
LE can be performed without manual intervention, however it is highly recommended that manual intervention (average time to complete: 4:59 min) and/or quality checking is performed.
Edit lesion segmentation
- Load lesion segmentation file on PD {session}_LEauto
- Use T1, PD/T2 to inform decision about lesion
- Reassign false negative (label 1 to label 2)
- Save as {session}_LEedit
- To ensure reproducibility, do not cut lesion mask masses unless necessary, if cutting choose the most narrow point
Manual tracing
- Large volumes of SH in ie. temporal poles or insula may not be included in LE mask because they are juxtacortical or based on intensity on PD
- In this case, manual tracing may be required to include these SH volumes
- The {session}_LEjux file may be used to inform manual tracing decision (as this file includes more false positives, and includes juxtacortical lesion of interest)
Note
- If your patient population has overt strokes that you wish to quantify, a manual tracing of the stroke penumbra must be completed and integrated into the lesion analysis
- You can use the closed polygon tool or paintbrush tool to create a stroke mask
- If you wish to quantify necrotic cores as well, a separate manual tracing must be completed
Common lesion and non-lesion variation
Due to anatomical variation and disease heterogeneity, lesions can manifest in many different ways on MRI. Neuroanatomical knowledge and experience using this pipeline are both important for accurate results. Below are examples of different lesion types and of things like Virchow-Robin (VR) spaces that may appear to be lesion. It is important to consider this when performing LE and/or FLEX.
Subcortical Hyperintensities (SH)
- SH appears hyperintense on both PD and T2, sometimes isointense on T1
- Easily identified large hyperintense regions covering white matter
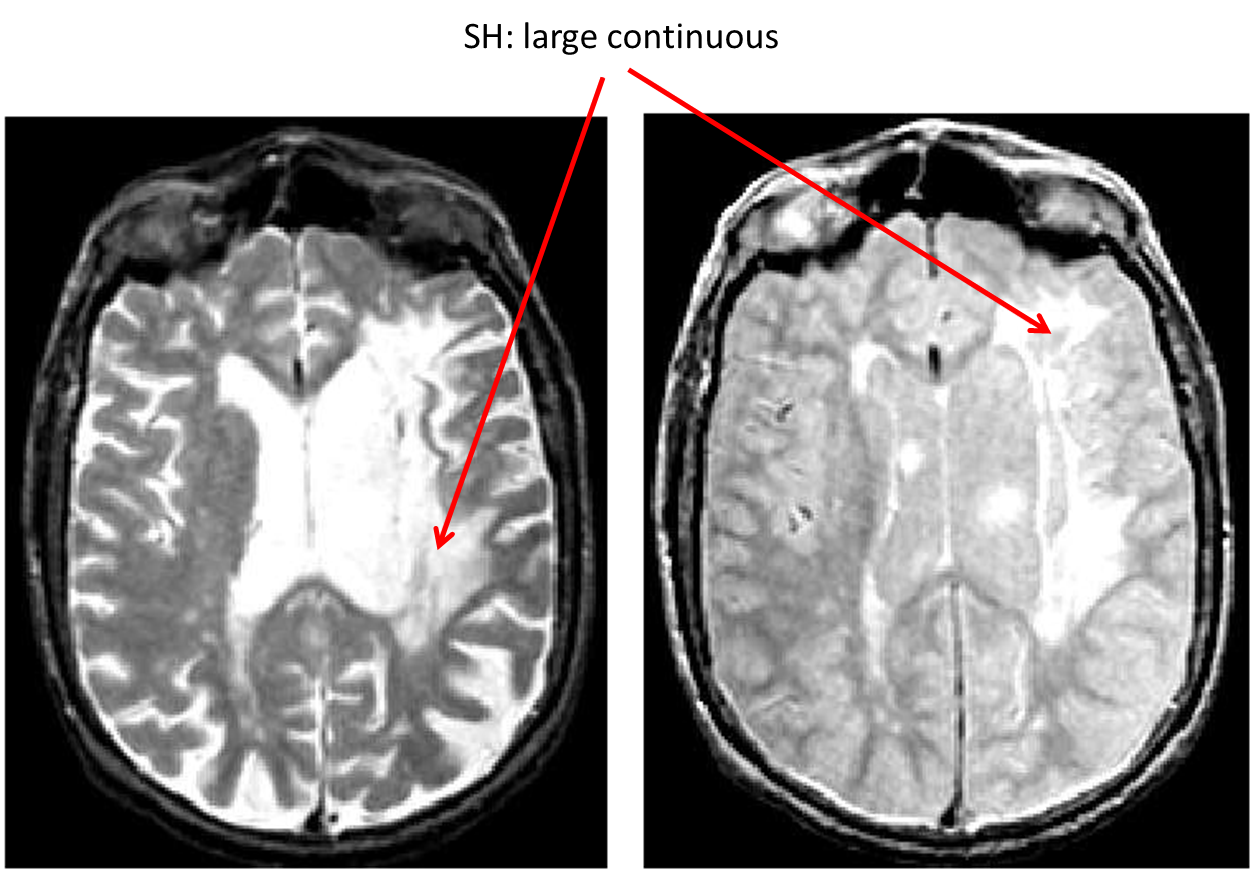
- SH also appears hyperintense on FLAIR
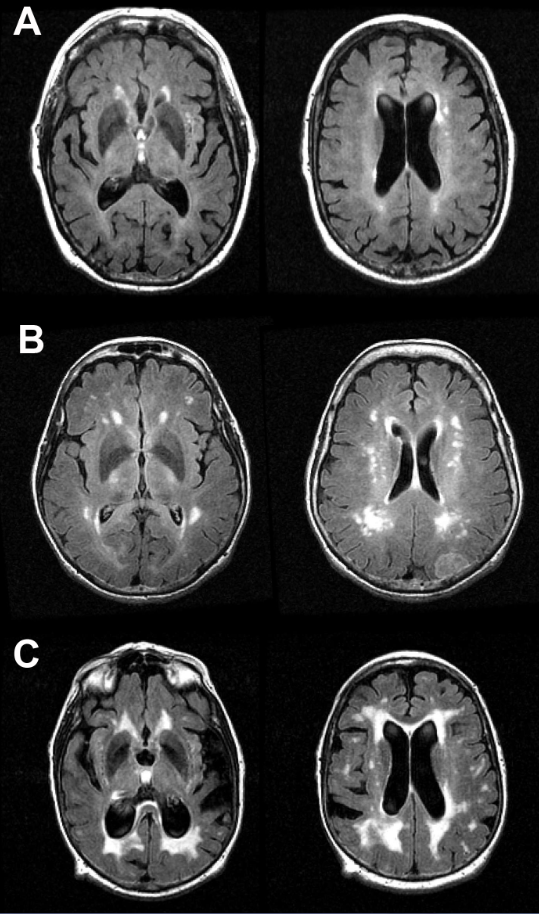
- Below are examples of A) mild SH, B) moderate SH, and C) severe SH on FLAIR images
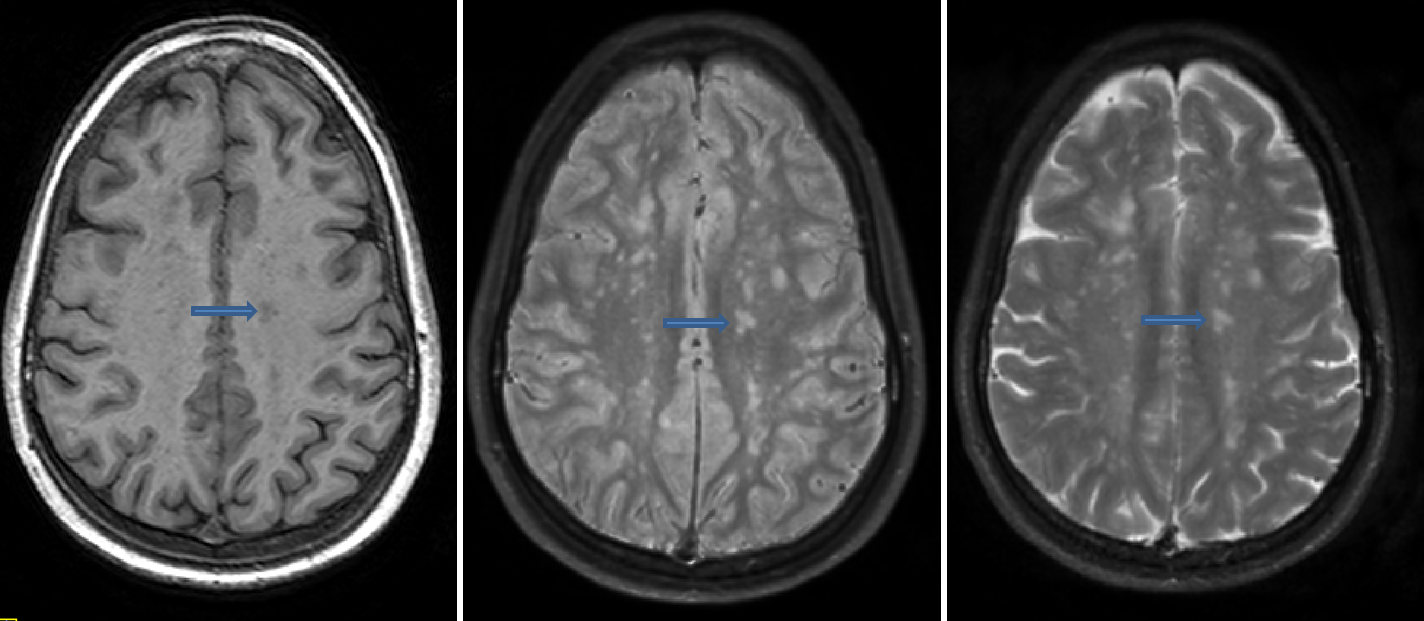
Discrete Subcortical Hyperintensity (SH)
- Characterized as multiple discrete hyperintensities on both PD and T2, and hypointense on T1
- Located in various regions throughout white matter
- Different from VR spaces, which are not hyperintense on PD
- Different from infarcts which are hypointense on T1
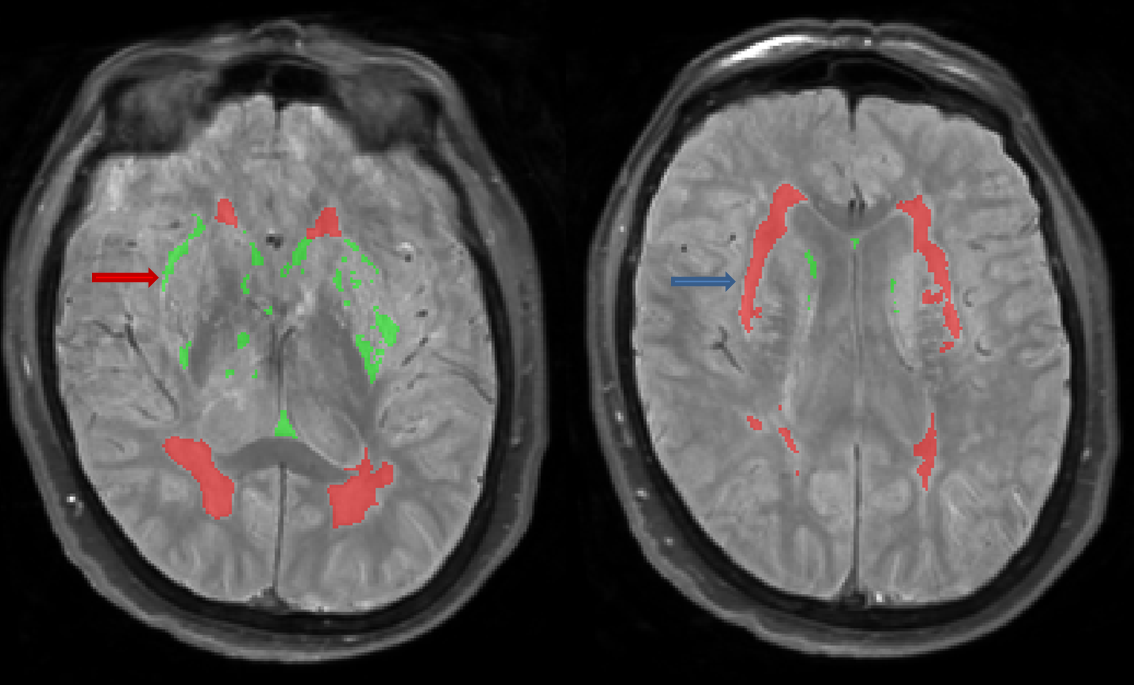
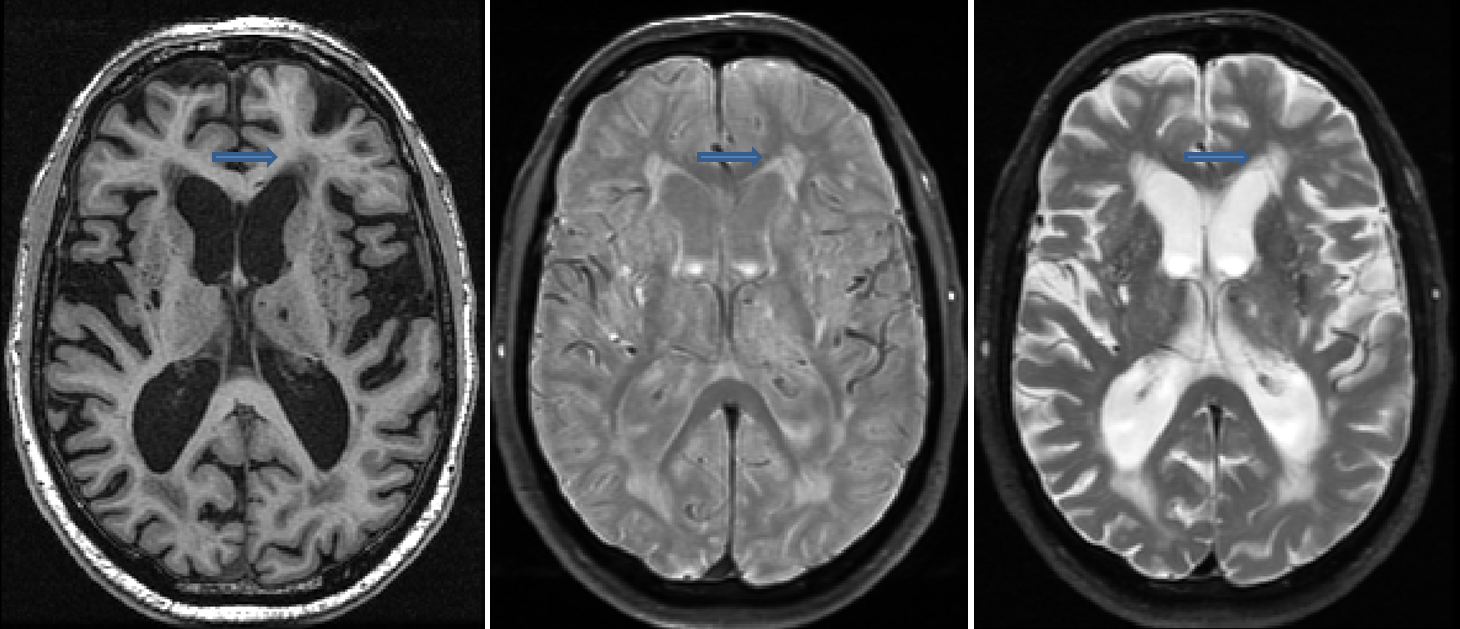
Periventricular Subcortical Hyperintensity (pvSH): smooth cap
- Hyperintense on both PD and T2, sometimes hypo/isointense on T1
- Characterized as a smooth hyperintense “cap” extending from the ventricles
- Easily identified on PD, difficult to distinguish from ventricular CSF on T2
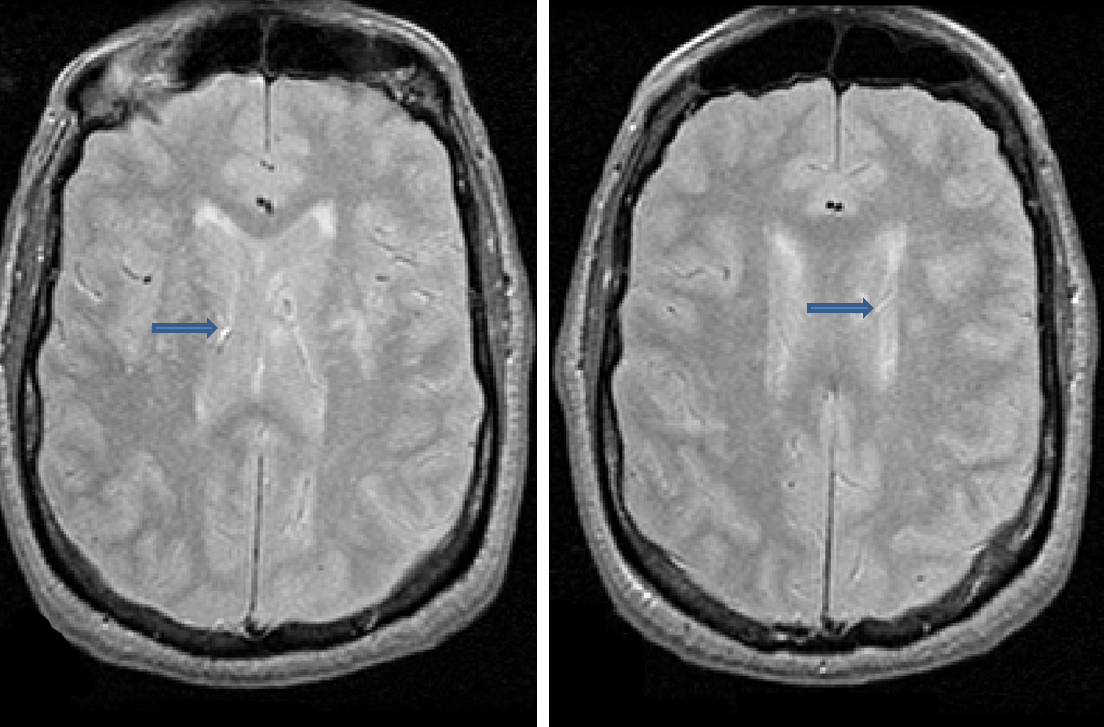
Vasculature
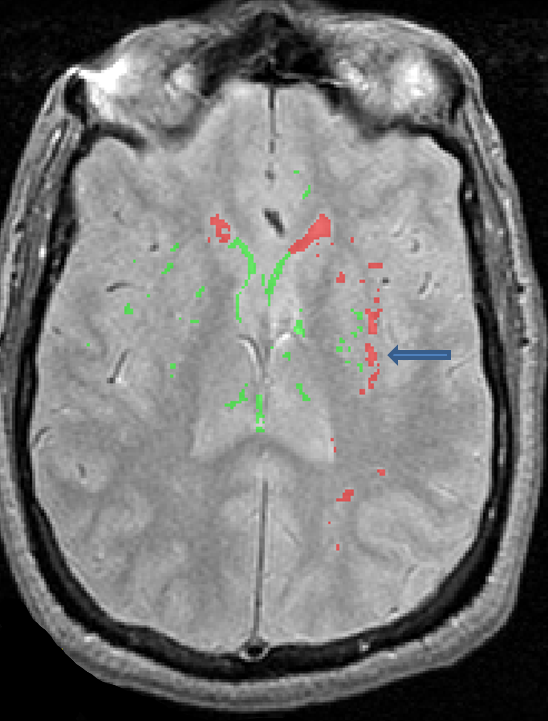
- Vasculature can appear bright on PD/T2 and may be mistaken for lesion
- It is important to look at multiple sequences and scroll through the slices of the image to determine whether a bright spot is lesion or vasculature
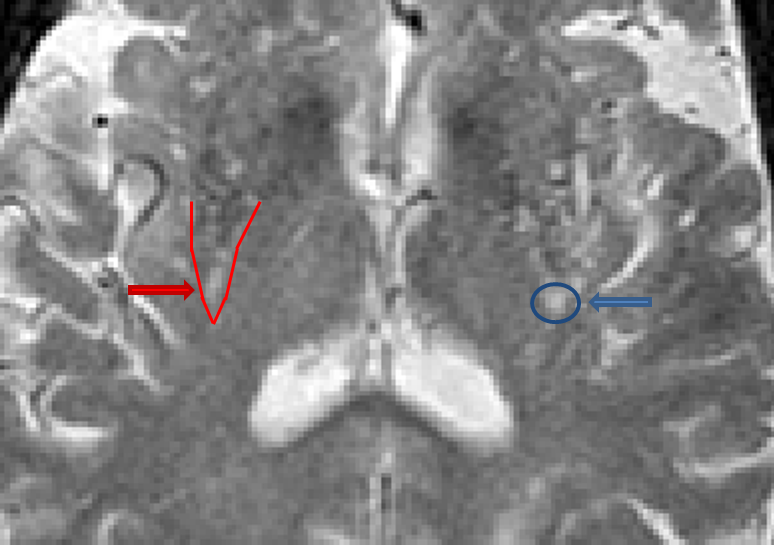
Virchow-Robin (VR) spaces
- Virchow-Robin (VR) spaces are enlarged perivascular spaces
- VR spaces are often located in and around the basal ganglia and thalamus regions, but are also found in superior slices above the ventricles as well
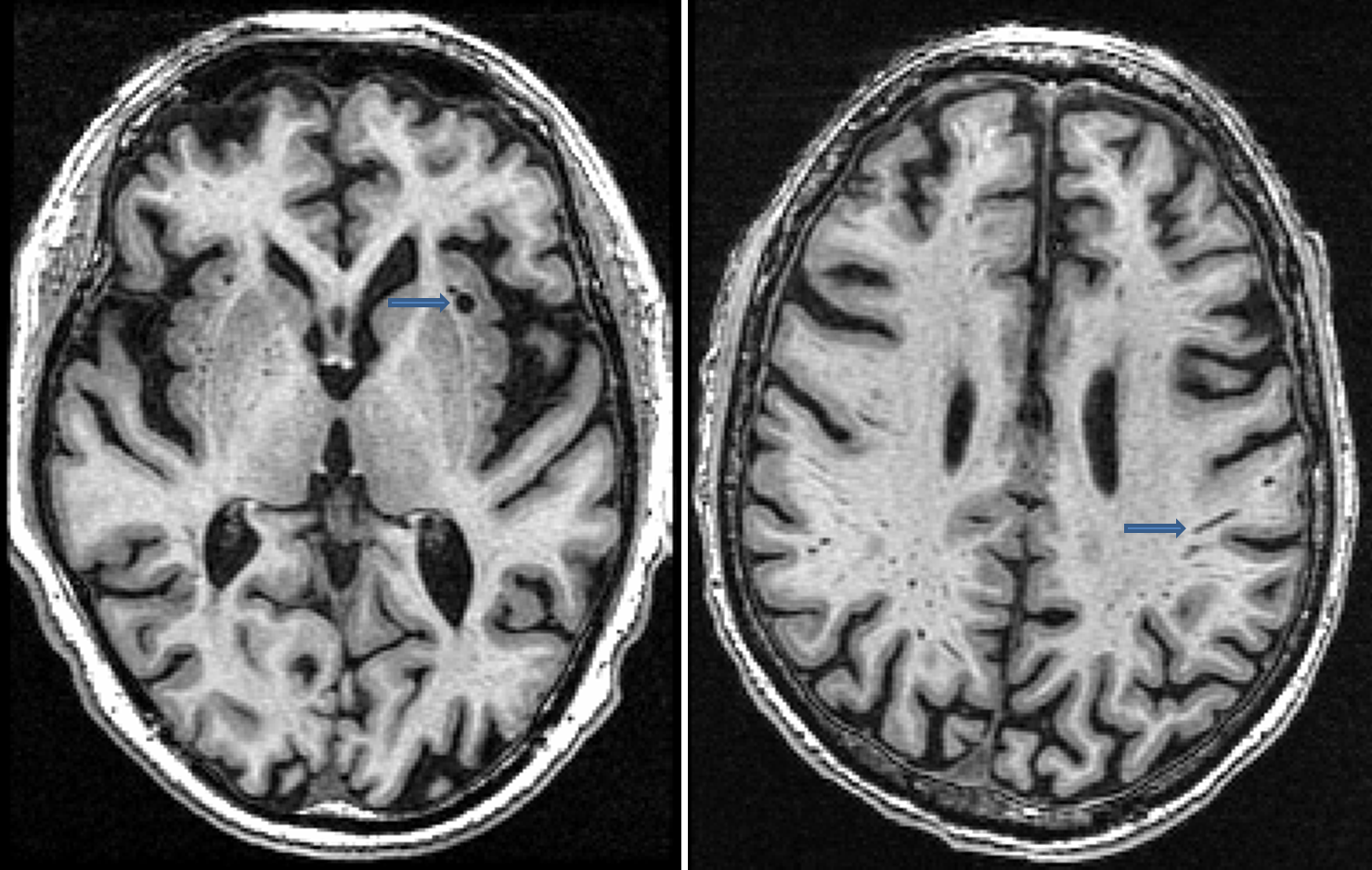
- VR spaces are characterized as hypointense on T1, isointense (or not visible) on PD and hyperintense on T2
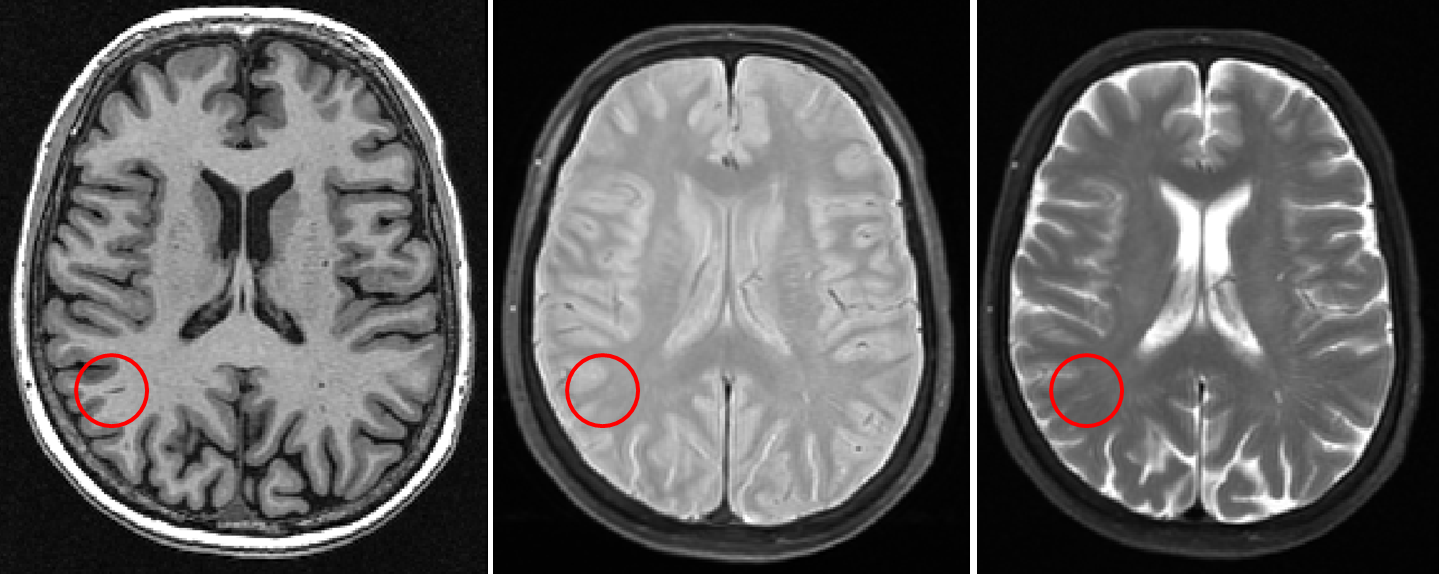
Corpus callosum
- Anterior medial portion of the corpus callosum may appear hyperintense
- This hyperintensity is actually within the ventricles and is not lesion
- Make sure to check the T1 as there are lesions found on the corpus callosum, however they are more oddly shaped and do not appear as a hyperintense “shadow” anterior to the corpus callosum
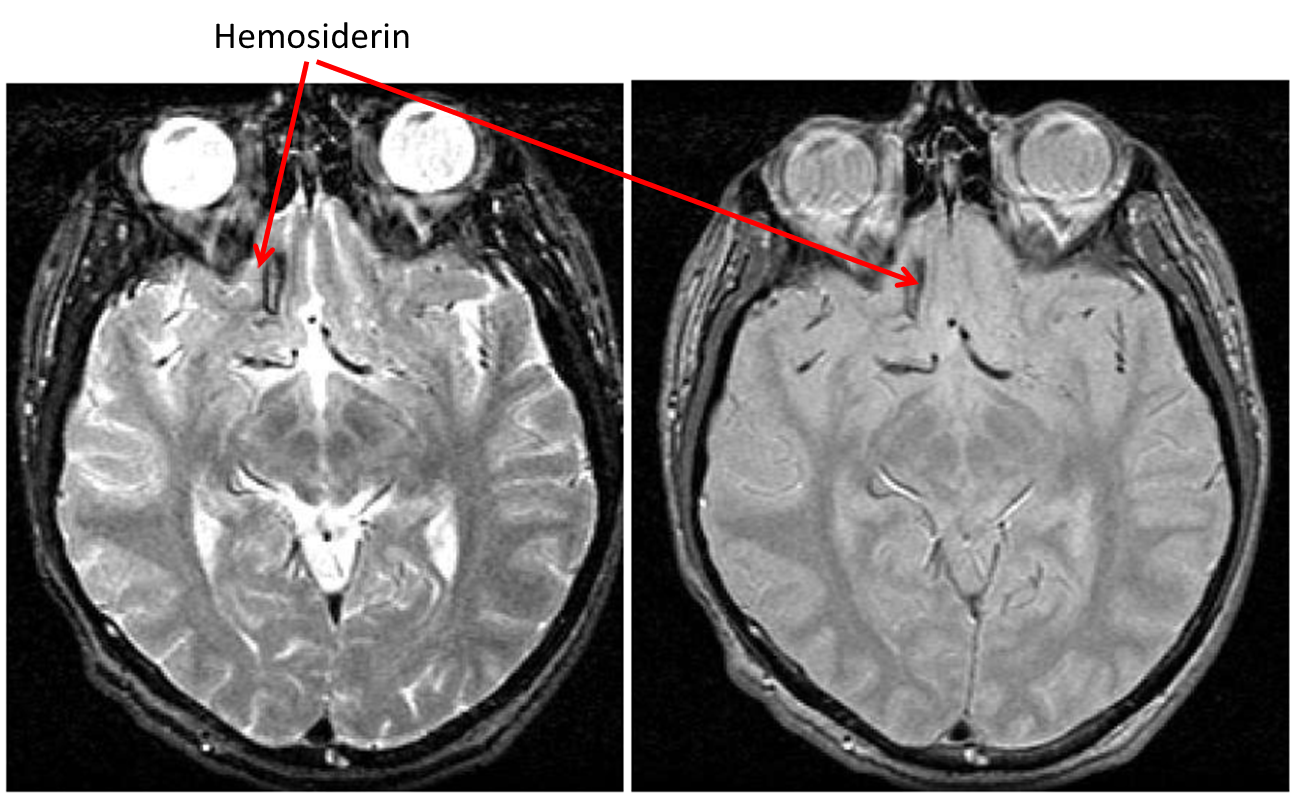
Traumatic Brain Injury (TBI): hemosiderin
- Hemosiderin is characterized as hypointense on both PD and T2
- These dark regions are signals from iron deposit left over from burst red blood cells post-trauma
- Unless you are looking for them, they are quite easy to miss as they are often located at the very edge of the brain as contusions
Files and parameters
Parameters
LE
| Parameter |
Value |
|---|
| minimum object size |
6 |
| -vcsf |
7 |
| -ct |
150 |
| -pdt |
0.10 |
| -t2t |
0.10 |
FLEX
| Parameter |
Value |
|---|
| -ct |
80 |
| -at |
80 |
| -nt |
4.5 |
Output files
| Filename |
Description |
|---|
| {session}_LEedit |
LE lesion mask after manual intervention / quality check |
| {session}_FLEXedit |
FLEX lesion mask after manual intervention / quality check |
Quality check
| Filename |
Description |
|---|
| {session}_LEauto |
Check for false positives and/or false negatives |
| {session}_FLEXauto |
Check for false positives and/or false negatives |